Engineering Computing Devices Using Bacteria Natural Signaling Systems
By Dr Daniel P. Martin, TSSG
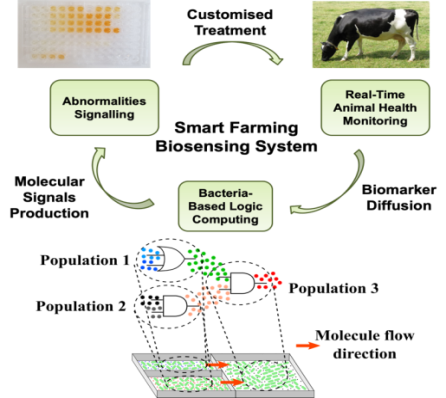
Recently, the advancements in animal microbiome research have provided insights into the role of bacteria signaling on the animal’s health. Bacteria can naturally produce molecules to coordinate behavior within a population upon detection of other signals or when in contact with surfaces. This natural communications system can be virulent and induce the animal’s immune system to counterattack, which result in infections.
On the other hand, the bacteria’s natural signaling system can be engineered to develop synthetic logic circuits that are suited for the design of specific biotechnological applications. For example, simple devices that perform logical operations, as well as oscillators, have already been developed by engineering both single cell and bacterial populations.
The integration of synthetic biology and molecular communications expertise expands the concept of artificial devices to enable the construction of robust bacteria-based computing systems that detect and process disease biomarkers to signal their presence in the animal’s gut. Furthermore, these same bacteria-based devices can output molecules or drugs to treat diseases originated from bacterial infections, such as metritis, bovine lameness, and mastitis.
Through VistaMilk, the investigation and modeling of more present and interconnected bacteria-based computing devices will enable the design of novel and customized therapeutics for chronic bacterial infections in cows. These engineering solutions will be based on the bacterial natural signaling processes and supported by the current and future communications and network systems.
Contact Daniel P.Martins : dpmartins@tssg.org
[/fusion_text]